By Dr. Tomislav Meštrović, MD, Ph.D. – Dec 13 2021 – News-Medical.net (link to article)
A recent review in the journal Life provided a detailed description of CRISPR-Cas-based diagnostic techniques to identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, but also emphasized the potential use of this technology towards developing effective molecular diagnostic platforms for any future pandemic.
The ongoing global pandemic of coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, has shown the importance of accurate diagnostic tests, especially for novel pathogens. The good news is that our diagnostic armamentarium is expanding, and it now also includes methods based on clustered regularly interspaced short palindromic repeats (CRISPR).
CRISPR and CRISPR-associated proteins (Cas) were initially found as components of the adaptive immune systems in bacteria and archaea (the latter being single-celled prokaryotic organisms distinct from bacteria), defending them against invading viruses by disintegrating foreign genetic elements.
Among all the different types of Cas nucleases identified thus far, Cas9 has been studied in detail and is commonly utilized for genome editing purposes. However, after the inaugural use of Cas9 in the first CRISPR-based diagnostic platforms, a sundry of Cas enzymes have been subsequently used to improve diagnostic approaches based on CRISPR.
More specifically, the distinctive collateral enzymatic activities of Cas12 and Cas13 have markedly reduced diagnostic times and costs, improving at the same time diagnostic accuracy and sensitivity. And this was what piqued the interest of Sangha Kwon and Ha Youn Shin from the Konkuk University in Seoul (Korea) in their recent review paper published in the journal Life.
Manifold advantages of CRISPR-Cas diagnostic platforms
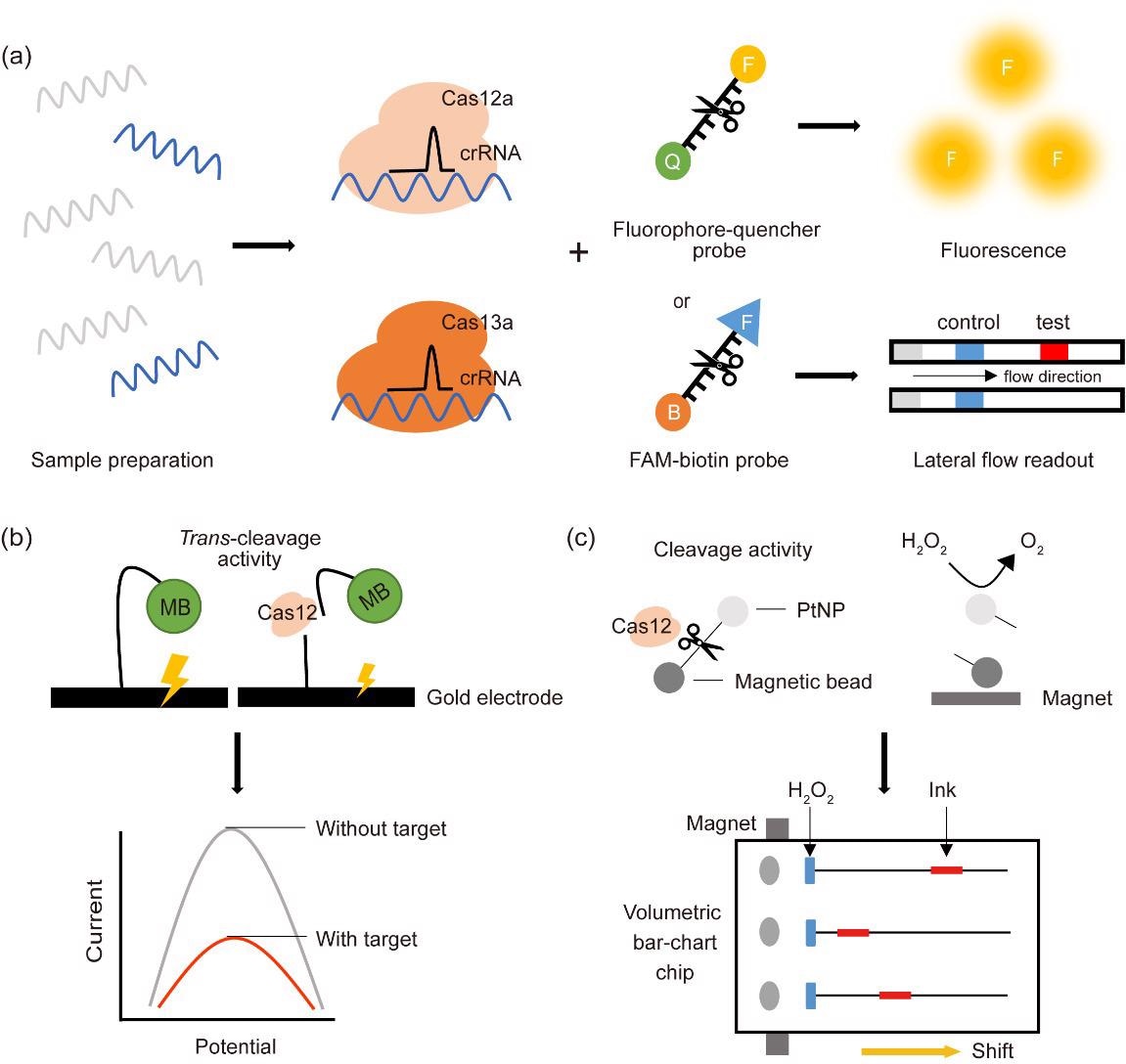
Some of the most pervasive CRISPR-Cas-based detection diagnostic platforms utilize Cas12 and Cas13 coupled with a fluorescent probe or lateral flow readout. Furthermore, there are trends in using an electrochemistry-based platform, which involves an affordable transduction element and a disposable sensor.
Such coupling of the unique collateral cleavage activities of Cas12 and Cas13 with different reporter systems has been essential in establishing a wide array of CRISPR-Cas-based molecular diagnostic platforms.
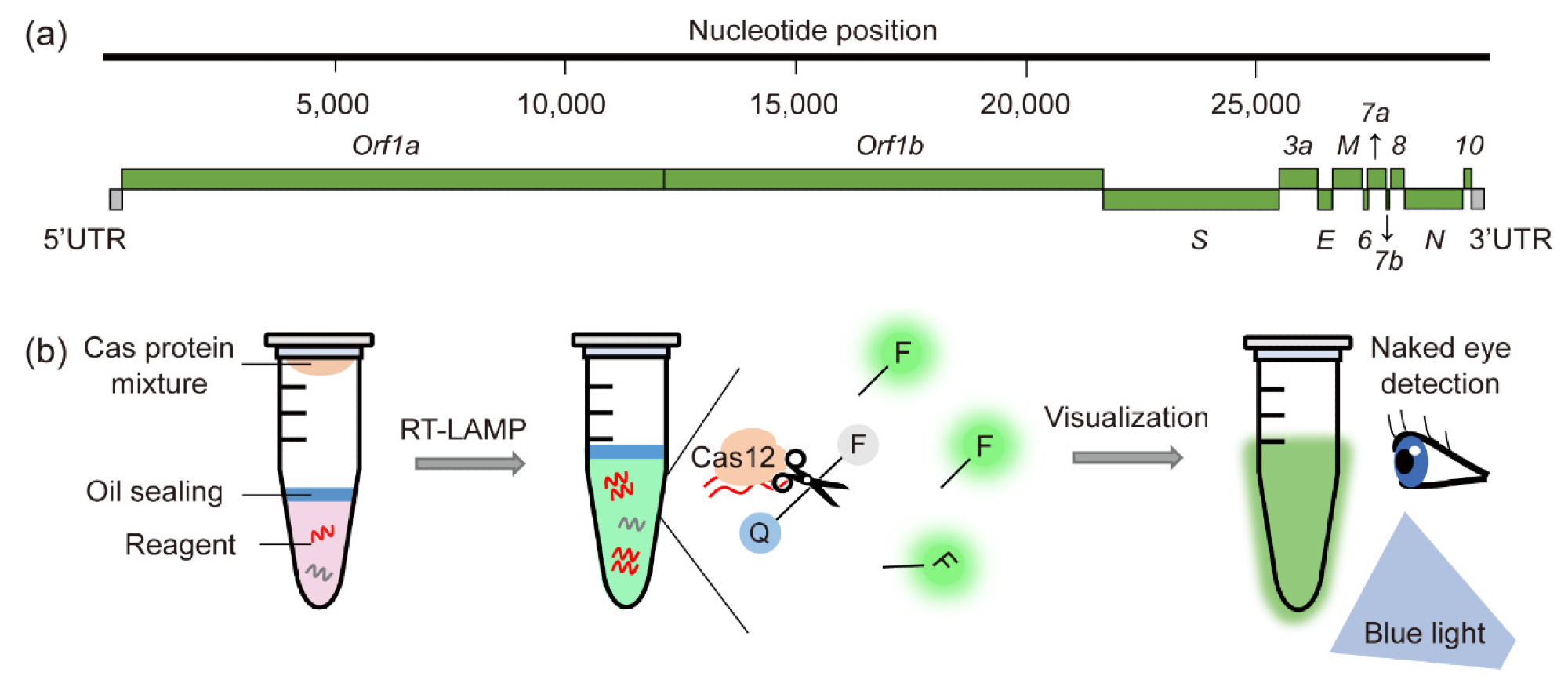
In comparison with the standard RT-PCR method for diagnosing COVID-19, CRISPR-Cas-based molecular diagnostic methods have improved sensitivity and target specificity. Furthermore, CRISPR-Cas-based systems are less expensive, more time-efficient, and much easier to perform in the field.
Limitations and off-target effects
Of course, such approaches also have certain limitations that may hinder their usability in quotidian clinical and diagnostic practice. For example, the nuclease activity of Cas proteins is actually highly dependent on the specific target sequence.
More specifically, this means that introducing random mutations into target sequences found in viral genomes will prevent pathogen detection. However, these limitations may be overcome by simultaneously using several guide RNAs targeting multiple pathogenic regions.
In addition, CRISPR-Cas systems have recognized off-target effects. This means there is a need to carefully design guide RNAs with off-target prediction software in order to circumvent the recognition of non-specific sequences, potentially leading to false-positive results.
Implications for future diagnostic approaches
As viral genomes are known to mutate rapidly, augmenting their ability to infect a wider variety of hosts and survive, we definitely need flexible diagnostic tools in the future that will target multiple regions simultaneously.
“The integration of artificial intelligence systems with disease diagnosis will enable patients to assess their diagnostic results through cell phone applications,” say study authors in this paper. “Furthermore, inexpensive portable systems could further contribute to the rapid diagnosis outside of hospitals and in developing countries,” they add.
In any case, CRISPR-Cas-based molecular methods may not be useful only during the COVID-19 pandemic, but we can view them as a promising strategy to prevent potential pandemics. Of course, further optimization is necessary to establish them as routine diagnostic tools.